Biological Sciences | Simulation, Data, Learning
Using Genome-Scaling Language Models to Reveal SARS-CoV-2 Evolutionary Dynamics
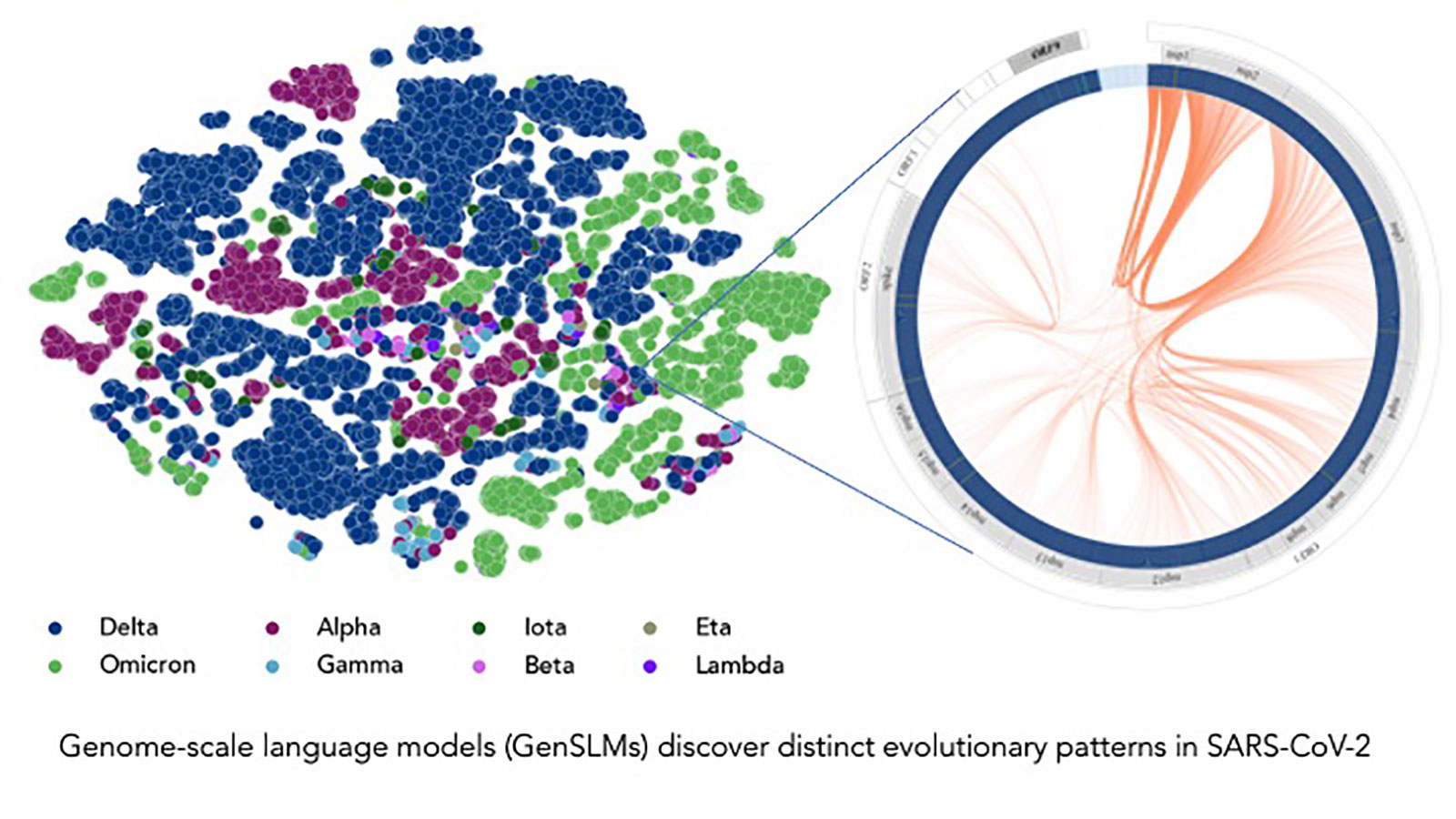
A research team led by Argonne National Laboratory scientists developed large-language models (LLMs)—models classically used to summarize, translate, and predict words in text—that can track virus evolution and mutation, and applied their work to SARS-CoV-2.
Challenge
Instead of reacting to viral mutations to subsequently identify variants of concern (VOCs), artificial intelligence (AI) and machine learning (ML) techniques can leverage deep sequencing data to identify mutations in viral proteins and characterize evolutionary patterns that help predict and describe future VOCs, potentially thereby transforming real-time pandemic monitoring. However, obtaining high-quality, global-scale genome datasets can be difficult. While sequence-based feature-extraction techniques followed by traditional ML approaches have demonstrated promise in the early identification of VOCs, they remain limited to sequence signatures of regions of interests in the genome. In this project, the researchers used LLMs in natural language processing tasks to develop global-scale, whole-genome surveillance tools, enabling them to characterize the evolutionary dynamics of SARS-CoV-2 while reconstructing the emergence of its variants.
Approach
The researchers relied on Argonne resources that included the Polaris supercomputer and Cerebras CS-2 AI platform. Utilizing 560 Polaris nodes—each of which includes four GPUs—helped the team scale their end-to-end workflow across the system in such a way that they were able to leverage the entire machine’s power to tackle complex calculations. The Cerebras platform enabled accelerated computation without compromises to accuracy.
Results
Through their efforts, the researchers generated the first genome-scale language model (GenSLM), enabling gene analysis as well as rapid identification of variants of VOCs. GenSLM is the first whole genome-scale foundation model that can be altered and applied to other prediction tasks similar to VOC identification.
Impact
This Gordon Bell Special Prize-winning work has the potential to lay the groundwork for a future pandemic observatory, enabling public health officials to respond quickly to virus variants, while also leading to protein-engineering applications or even the modeling of entire organisms.